Small cell lung cancer (SCLC) is a high-grade neuroendocrine cancer that is among the most lethal solid tumor malignancies, with a 5-year overall survival of less than 5%. There are no effective therapies available for treating SCLC, and treatment outcomes have remained virtually unchanged for the past thirty years. Our lab uses genetically engineered mouse models of SCLC to better understand two important questions: 1) what are the mechanisms of SCLC progression from early pre-cancerous tumors to highly metastatic cancers, and 2) why do these tumors fail standard chemotherapy treatments?
Mice are engineered to lose the genes encoding the key tumor suppressors p53 and Rb in the precursor lung cells that give rise to SCLC. These animals develop highly metastatic, lethal, SCLC tumors. We have applied modern genomics and sophisticated live-animal imaging methods to show that tumors in the mouse model acquire additional “driver” genetic alterations that contribute to the aggressive behavior of SCLC (Dooley et al., 2011, McFadden et al., 2014). In addition, these studies have suggested that tumors may spread (metastasize) earlier and in more complex patterns than previously hypothesized (McFadden et al., 2014).
There are two major subpopulations within SCLC tumors, one is neuroendocrine-like (NE), while the other is non-neuroendocrine-like (non-NE). Over the past few years, we have fully characterized these two cell types, both functionally and by gene expression profiling. We found that these two cell types collaborate at multiple stages of cancer metastasis, and dissected out the mechanisms underlying such collaboration. We also identified molecular markers for these two cell types; using these markers, we were able to demonstrate the co-localization of these two cell types in autochthonous SCLC tumors.
Recent genomic sequencing studies of human small cell lung cancer (SCLC) tumors have identified recurrent mutations in multiple genes, most of which have not been functionally characterized in SCLC. We have developed a CRISPR-Cas9-based system to generate loss-of-function mutations in developing tumors in vivo, which allows us to rapidly model and functionally characterize these candidate driver mutations in our mouse model of SCLC. We have crossed a Cre-inducible Cas9 allele into the p53fl/fl; Rbfl/fl model of SCLC. Using this model, we can carry out somatic gene editing specifically in tumor cells via viral delivery of Cre recombinase and target sgRNAs. Proof-of-principle studies involving deletion of known tumor suppressor genes in SCLC demonstrated acceleration of tumor progression in these animals compared with control cohorts, validating our approach. Our current work extends these studies using methods to identify and study circulating cancer cells, pre-clinical studies using targeted and combination therapies, and genome-engineering approaches to identify, and target, additional drivers of SCLC progression and treatment resistance.
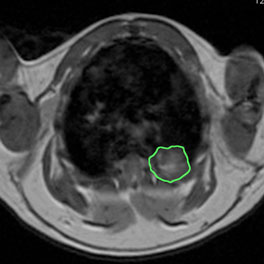
Thoracic MRI image of a p53flox/flox; Rb1flox/flox mouse 14 months after intratracheal infection with adenovirus expressing Cre recombinase. Tumor is outlined in green.

